Workflow & Overview
SegMantX is organized into modules. This is a graphical abstract for the suggested workflow:
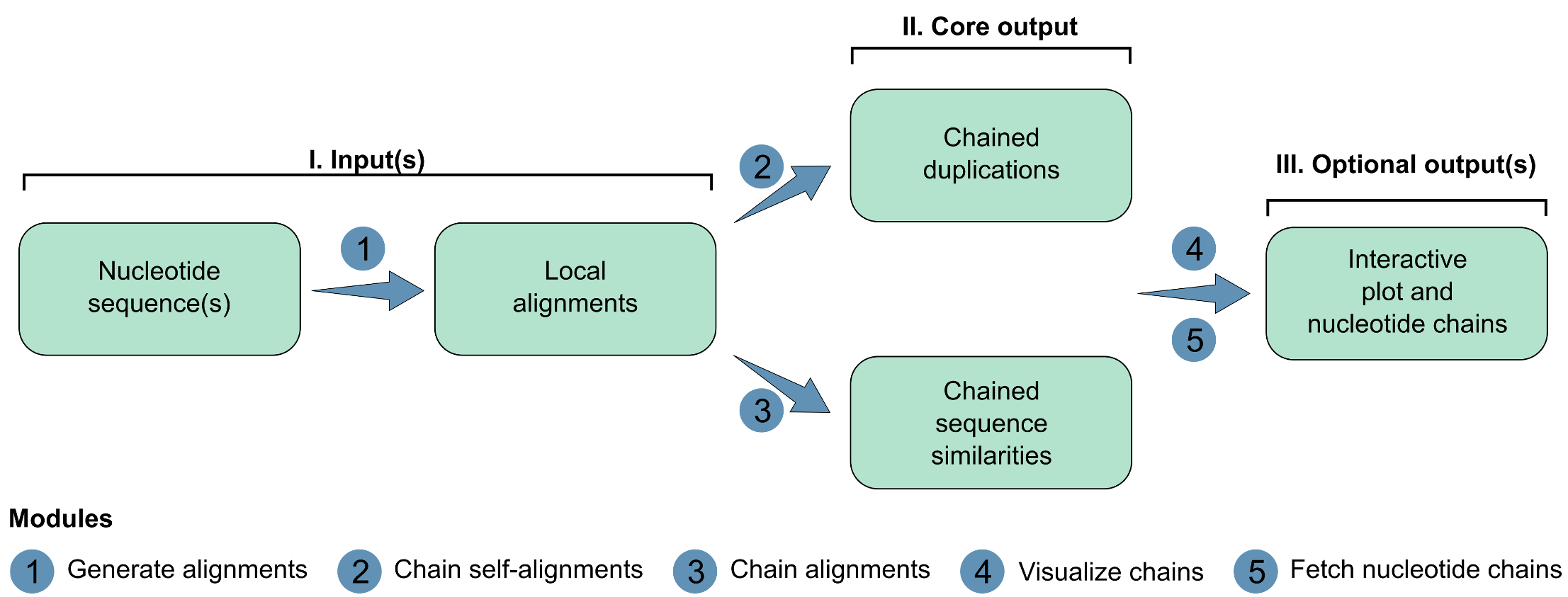
- Generate alignments: processes nucleotide sequence(s) to compute local alignments, optionally formatting them for further analysis.
- Chain self-alignments: Chains local alignments from self-sequence alignment (e.g., towards duplication detection).
- Chaing alignments: Chains local alignments between two sequences (e.g., towards sequence comparisons).
- Visualize chains: Generates a segmentplot (i.e., segments of chaining results) to visualize yielded chains for a sequence (self-alignment) or two sequences (alignment).
- Fetch nucleotide chains: Extracts yielded chains as nucleotide sequences and saves them as fasta file.
